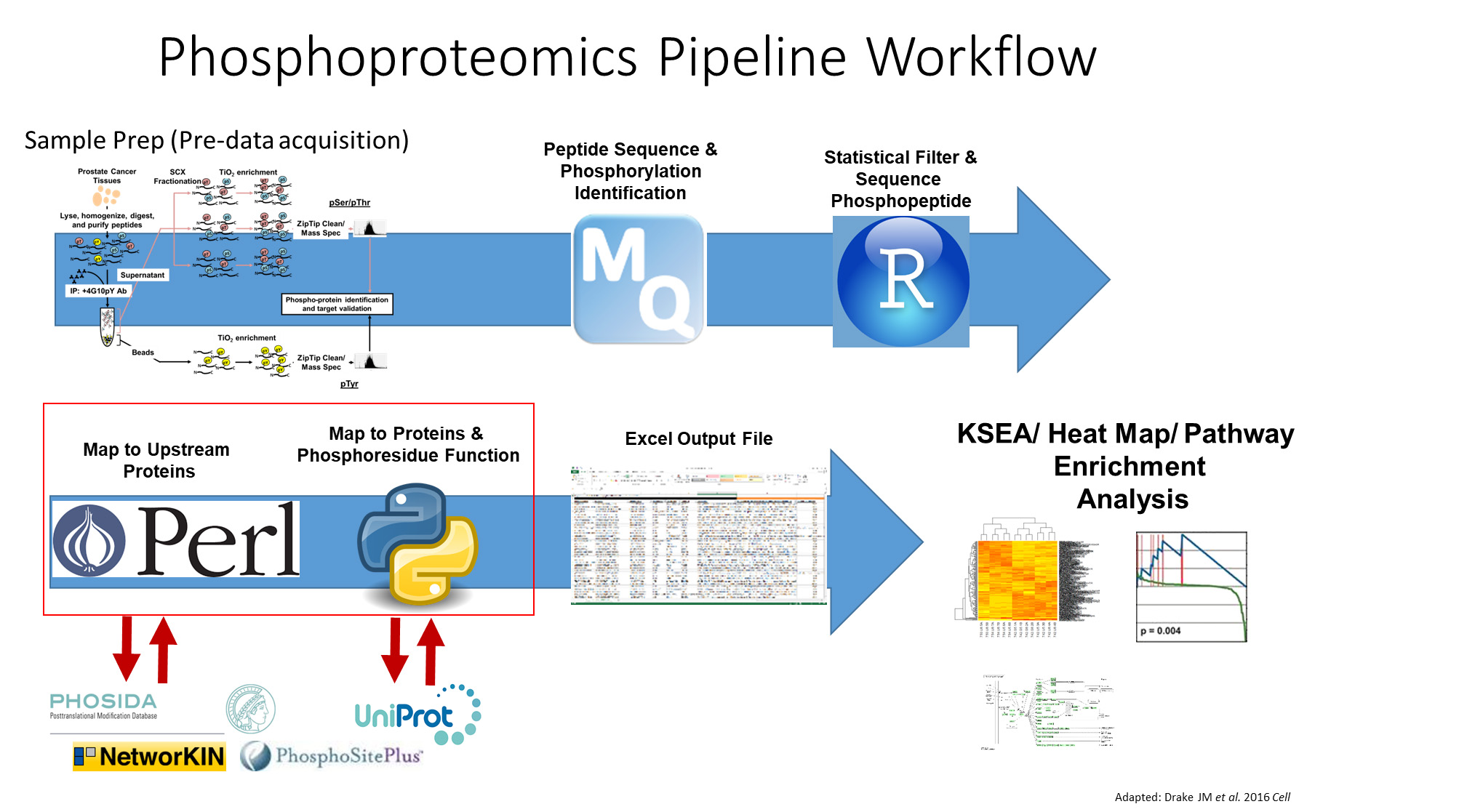
Many researchers struggle with proteomics large data analysis and how to implement these analyses. Furthermore, there is a large demand for free and accessible platforms with straightforward workflows. Currently, all proteomics data processing and analysis platforms available require a high financial investment while offering limited return, such as a few graphed results and rudimentary quality control analysis. Within such frameworks, data analyses require that investigators implement multiple steps, and manual manipulation of the data can incur a high probability of errors; it is inefficient and time consuming.
Assistant Professor Justin Drake (Pharmacology; Masonic Cancer Center; MSI PI) and his team have developed a platform that provides a straightforward workflow that not only automates data manipulation and searches but also provides statistical analysis that includes comprehensive quality control outputs, downstream output results such as kinase analysis, pathway construction, and phosphosite analysis with graphs and heat maps. The current project, which recently received a UMII Seed Grant, intends to utilize the Galaxy instance hosted by MSI to make the platform accessible to all University of Minnesota researchers.
UMII Seed Grant funds are intended to promote, catalyze, accelerate and advance U of M-based informatics research in areas related to the MnDRIVE initiative, so that U of M faculty and staff are well prepared to compete for longer term external funding opportunities. This project falls under several research areas of the MnDRIVE initiative:
- Global Food Ventures
- Advancing Industry, Conserving Our Environment
- Discoveries and Treatments for Brain Conditions
- Cancer Clinical Trials
Research Computing partners:
- University of Minnesota Informatics Institute
- Minnesota Supercomputing Institute