Tunneling nanotubes (TNTs) are long, thin, spontaneously forming actin-based cellular extensions that occur in a variety of cell types, including inflammatory cells, neurons, and malignant cells. TNTs form direct connections that serve as conduits for intercellular transport of a variety of cellular cargo and contents, including but not limited to lipophilic vesicles, Golgi vesicles, and even mitochondria. MSI PI Emil Lou (associate professor, Medicine) studies TNTs and his research group was the first to demonstrate, using confocal microscopy, the presence of nanotubes in intact malignant tumors. They have since discovered TNTs in several different forms of cancer. Professor Lou and his collaborator, MSI PI Subbaya Subramanian (associate professor, Surgery), wrote a chapter on TNTs for the textbook Intercellular Communication in Cancer: Lou E., Subramanian S. (2015) Tunneling Nanotubes: Intercellular Conduits for Direct Cell-to-Cell Communication in Cancer. In: Kandouz M. (ed.) Intercellular Communication in Cancer. Springer, Dordrecht.
It is a time-consuming process to manually identify tunneling nanotubes between cells in images. A collaborative team has developed a machine-learning algorithm to automate this process. A large team of researchers collaborated to complete this project:
- Dr. Thomas Pengo (Director, University of Minnesota Informatics Institute) has worked with the Lou Lab on this project since 2015. Three students in the lab, Snider Desir, William Sperduto, Akshat Sarkari), provided example data and tested early versions of the software. Dr. Pengo has coordinated the collaboration between UMII, MSI, and the Lou Lab.
- When conventional image analysis methods were not working well, the lab applied for a UMII Updraft (now Small UMII Seed Grant (link: https://research.umn.edu/units/umii/funding/overview) to ask the Scientific Computing Solutions group at MSI to collaborate to develop the Deep Learning algorithm.
- Sophie Korenfeld, an undergraduate in Dr. Lou's lab, provided crucial manual annotations to train the algorithm, without which none of this would be possible.
- Dr. Ben Lynch in MSI's Scientific Computing Solutions group was the developer of the first version of the Deep Learning algorithm
- Ben Johnson, an undergraduate student researcher at UMII, worked with Dr. Pengo to fine-tune the metaparameters of the algorithm, with some key successes. Mr. Johnson has graduated and is working in industry.
- Charlie Bradley, an undergraduate student researcher at UMII, continued the project. He worked with MSI’s Dr. Ham Lam (Scientific Computing Solutions). Using GPUs on MSI’s Mangi supercomputing cluster, they were able to port the code, improve it, and train it. Mr. Bradley graduated with a Computer Science bachelor’s degree in 2020.
- Support from other members of the Lou Lab also contributed to the project.
- Mark Sanders and Guillermo Marques at the University Imaging Centers helped to improve the image-acquisition part of the project.
Research Computing partners:
- Minnesota Supercomputing Institute
- UM Informatics Institute
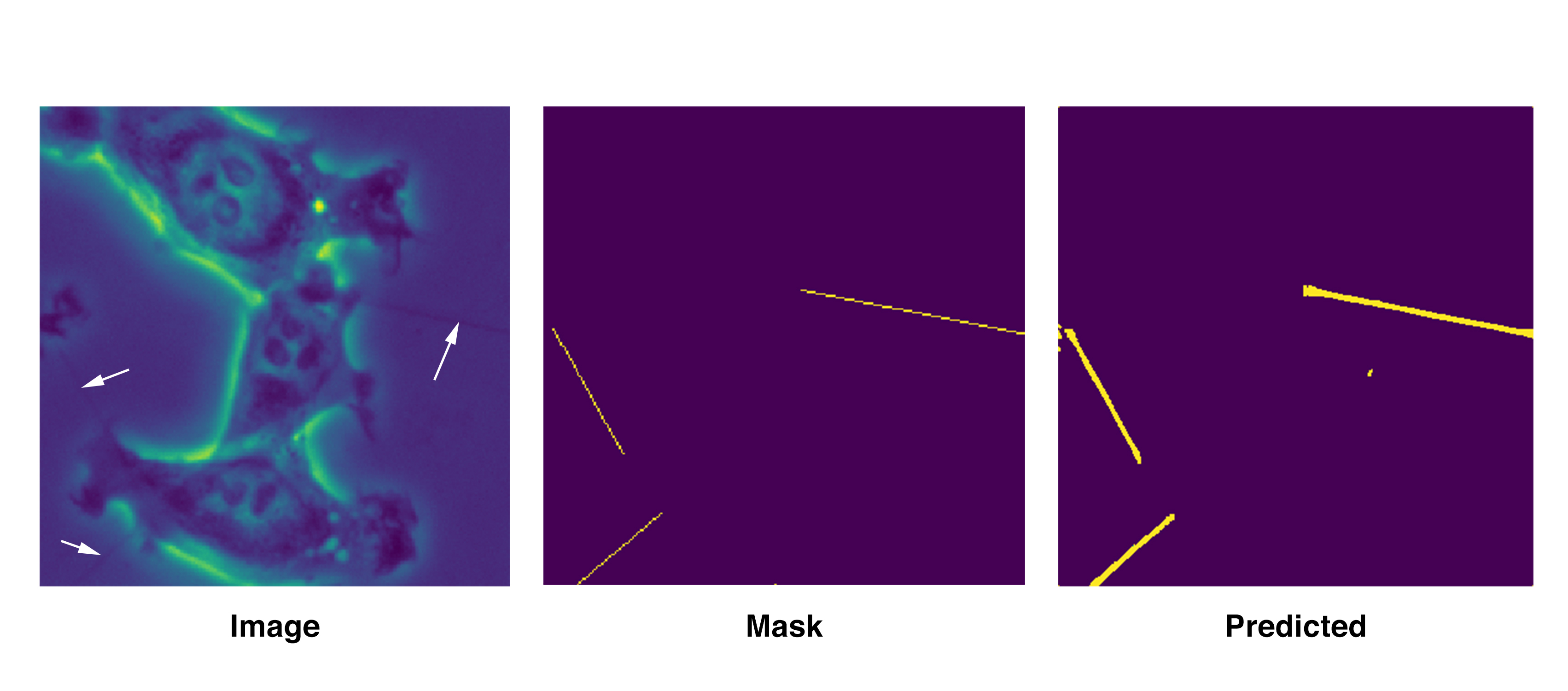
Image description:
Left: A test image for the UNet deep learning algorithm showing the long and thin tunneling nanotubes for intercellular communication. The tubes (pointed out by arrows) are very faintly visible on this image.
Center: A mask image that corresponds to the test image serves as the correct “label” showing three thin nanotubes in the test image.
Right: A UNet generated image showing three predicted tunneling nanotubes after seeing the test image.