The zebra mussel (Dreissena polymorpha) is a freshwater invasive species that has caused billions of dollars of damage to waterways in Europe and North America. Economic effects include damage to the power-generation industry and to recreation and tourism. The invasive mussels also crowd out native species, destroy those species’ food sources as they remove large quantities of phytoplankton in the water, and cause dramatic changes to the food web.
In order to better understand and control this species, a group of researchers including MSI Principal Investigators and their group members, Research Computing staff members, and colleagues at the University of Toronto, Phase Genomics, and the National Human Genome Research Institute are studying the D. polymorpha genome. In a paper published recently in the journal G3, the researchers used long-read sequencing and scaffolding technologies to generate a chromosome-scale assembly of the zebra mussel genome. This is the highest quality genome assembly of a mollusk to date. The genome also fills in a large gap in the tree of life, since zebra mussels are more than 400 million years diverged from their nearest sequenced relative. This is roughly the same amount of evolutionary divergence as that between humans and manta rays. They also have the largest mitochondrial genome of any animal studied to date, although the reason for this unusual mitochondrial genome structure is unknown.
The chromosome-level assembly used in this study is a powerful new tool for basic and applied research on this and related invasive bivalves. The genome provides insights into the processes that have made zebra mussels so successful as an invasive species. The researchers identified genes associated with some of them, such as shell formation, byssal thread attachment fibers (these help the mussel attach to surfaces and to spread between water bodies), and responses to thermal stress. These genes and others are targets for development of control technologies.
The paper can be read on the website of the journal G3: Michael A McCartney, Benjamin Auch, Thomas Kono, Sophie Mallez, Ying Zhang, Angelico Obille, Aaron Becker, Juan E. Abrahante, John Garbe, Jonathan P. Badalamenti, Adam Herman, Hayley Mangelson, Ivan Liachko, Shawn Sullivan, Eli D. Sone, Sergey Koren, Kevin A.T. Silverstein, Kenneth B. Beckman, Daryl M. Gohl. The Genome of the Zebra Mussel, Dreissena polymorpha: A Resource for Invasive Species Research. 2022. G3 12(2). doi: 10.1093/g3journal/jkab423.
Research Computing partners:
- University of Minnesota Informatics Institute
- Minnesota Supercomputing Institute
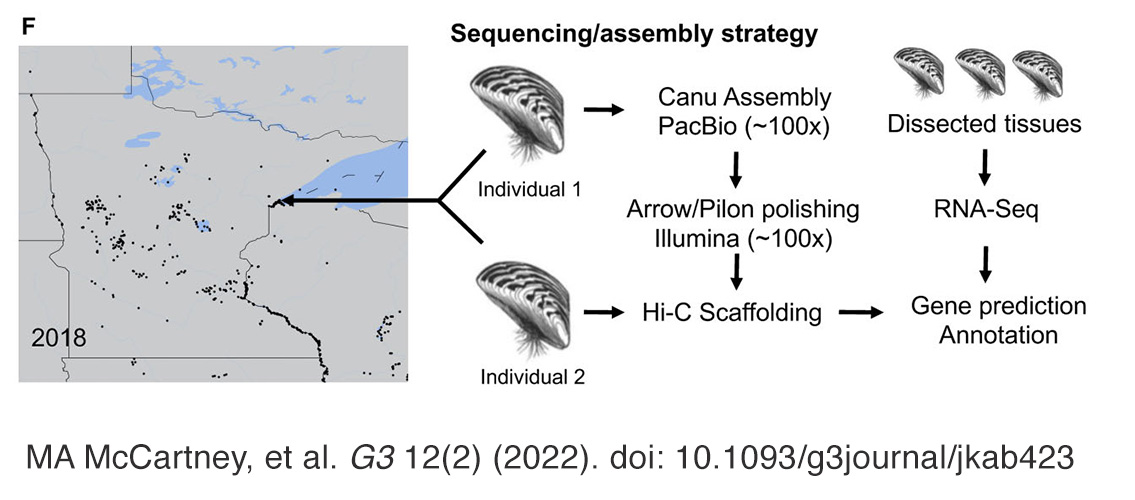
Image description: Left: Map showing the extent of zebra mussel infestation in Minnesota lakes as of 2018 and depicting the location where the specimens for genome sequencing and scaffolding were collected. Right: Summary of the sequencing and annotation strategy. Image and description: MA McCartney, et al. G3 12(2) (2022). doi: 10.1093/g3journal/jkab423