Diseases caused by plant pathogens reduce the global food crop by 40% annually. However, the bacteria and fungi that live in the soil (soil microbes) can produce an astounding diversity of antibiotic compounds that help crops and plants combat and suppress pathogens. In contrast to chemical-based pesticides, this antibiotic-mediated disease suppression by soil microbes presents a promising tool towards more sustainable disease control and yield stability, however, we still don’t understand much about the factors that influence the soil microbiome and antibiotic production. Particularly in agricultural contexts, we lack insight into the ways in which long-term farm practices, such as heavy fertilizer inputs, may alter the abundance and expression of antibiotics within the soil-microbiome. Thus, our ability to develop and manage disease-suppressive soils has remained severely limited.
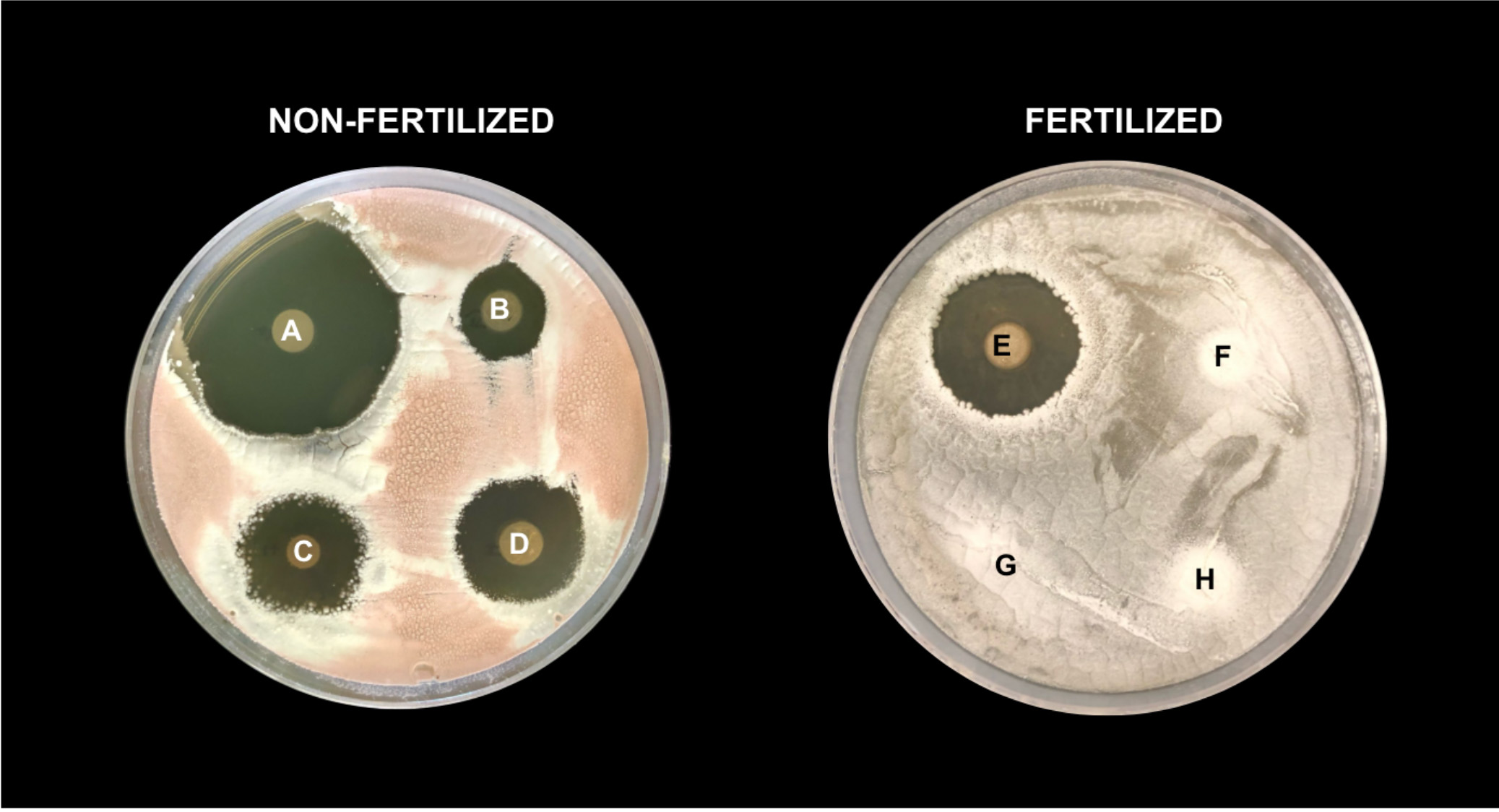
Previous research has shown that antibiotic production is significantly reduced among soil bacteria from high-nitrogen (fertilized) versus low-nitrogen (non-fertilized) fields. However, we do not understand whether the differences in antibiotic production among bacteria from fertilized conditions reflects a decrease in the number of antibiotics being expressed (suggesting that fertilizer application maybe “turning off” antibiotic production) or instead reflects soil bacteria with fewer antibiotic production genes (suggesting fertilizer application is resulting in a genomic loss of antibiotic production). While a genomic loss in antibiotic genes between fertilized vs. non-fertilized bacteria would suggest a fundamental shift in disease-suppressive potential of the microbiome, observed differences in antibiotic gene expression could lead to soil management strategies to help “turn on” antibiotic production in the soil’s existing microbiome. Understanding how fertilizer application shapes antibiotic production within soil microbiomes will be a critically important step towards optimizing disease suppressive soils and providing farmers with more wholistic disease management strategies.
PhD student Molly Kuhs (Ecology, Evolution, and Behavior), in a project called “Does Long-Term Soil Nutrient Management Alter the Disease-Suppressive Capacity of Soil Microbes?”, is utilizing multiple bioinformatic approaches, including whole-genome and transcriptomic analyses, to examine two populations of soil microbes that have been under distinct long-term fertilized and non-fertilized conditions. The project will evaluate if fertilizer application results in changes in the antibiotic gene expression, or losses in antibiotic genes. Together, the resulting data will provide a deeper understanding of how fertilizer application shapes the disease-suppressive capacity of soil microbiomes and will provide crucial context for building long-term management strategies for reducing disease, sustainably supporting crop health, and generating yield stability across both local and global levels.
Some funding for this project was provided by a 2022 University of Minnesota Informatics Institute MnDRIVE PhD Graduate Assistantship. The UMII MnDRIVE Graduate Assistantship program supports UMN PhD candidates pursuing research at the intersection of informatics and any of the five MnDRIVE areas:
- Robotics, Sensors and Advanced Manufacturing
- Global Food Ventures
- Advancing Industry, Conserving Our Environment
- Discoveries and Treatments for Brain Conditions
- Cancer Clinical Trials
This project is part of the Global Food Ventures MnDRIVE area.
Research Computing partners:
- University of Minnesota Informatics Institute
Complete list of 2022 UMII MnDRIVE PhD Graduate Assistantships.
Image description: Petri dishes containing dotted soil bacteria (labeled A-H) covered with a pathogen. Clear “no-growth” rings around each bacteria indicate antibiotic production from the dotted bacteria that is inhibiting the growth of the pathogen. Examples of bacteria taken from long-term non-fertilized (left) and fertilized (right) fields showcase how fertilizer application leads to variation in antibiotic production and ability to inhibit the growth of pathogens among members of the soil microbiome.