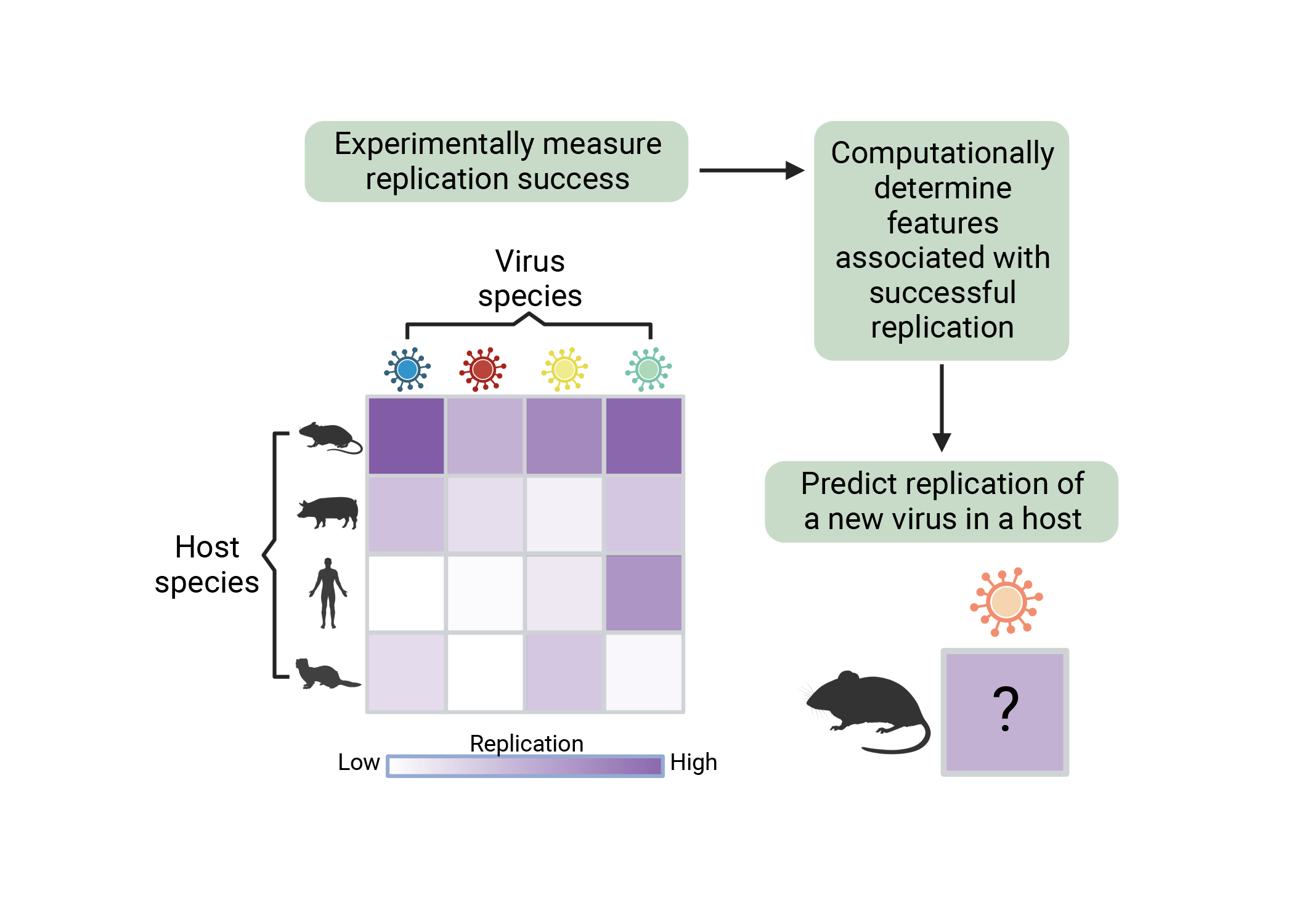
An estimated 1.6 million viruses are capable of infecting mammals and birds, and the risk of virus spillover from one host into another is increasing with climate change and expanding human populations. However, determining which viruses have the potential to infect humans and drive novel pandemics is largely unknown.
Associate Professor Ryan Langlois (Microbiology and Immunology; MSI PI) and Associate Professor Kimberly VanderWaal (Veterinary Population Medicine; MSI PI) are working on a project called “Computationally predicting viruses with cross-species replication potential,” that seeks to address this large gap in knowledge by measuring replication of viruses in fibroblast cell lines that represent the diversity of Mammalia. Using statistical approaches including machine learning, the project goal is to determine whichvirus and host characteristics are most associated with successful viral replicationacross mammalian species. Taken together, this work will generate the largest andmost diverse experimental virus-host data set ever assembled. The host and virusfeatures that are most associated with successful replication can be used to assesscross-species transmission risk of novel viruses and help prevent future pandemics.
This project recently received a Research Computing Seed Grant. RC Seed Grant funds promote, catalyze, accelerate and advance U of M-based informatics research in areas related to the MnDRIVE initiative, so that U of M faculty and staff are well prepared to compete for longer term external funding opportunities. This Seed Grant falls under the Global Food research area of the MnDRIVE initiative. The project has also received in-kind support from the University of Minnesota Institute for Infectious Diseases.
As of September 2023, the RC Seed Grant programs have been revised into the DSI Seed Grant programs. DSI Seed Grants include many of the same goals as the old program, with a new emphasis on data science. See complete information about DSI Seed Grants, including application deadlines.