Hydrocephalus is a condition described as an excess of cerebrospinal fluid (CSF) in the ventricular system in the brain, caused by either impaired absorption or overproduction. It affects 1 in 500 children, and in 2003, despite accounting for a small percentage of pediatric hospital admissions, hydrocephalus accounted for a disproportionate number of hospital days and charges. Despite its multitude of causes, it is classically and contemporarily still considered a surgically correctable problem. The mainstay of treatment focuses on CSF diversion, either shunting or other means. Although its economic cost to the health system is quantifiable and profound, with significant financial and human impact on patients and families as well, clinical outcomes for pediatric hydrocephalus have not seen meaningful improvement. The pediatric neurosurgery community agrees that there must be understanding of the fundamentals of CSF hydrodynamics, particularly at the developmental level, which would be captured predominantly most plainly by neuroimaging. A rapid and accurate assessment of pediatric ventricles is needed to provide a quantitative understanding of ventricular development throughout childhood, particularly in the setting of pathology.
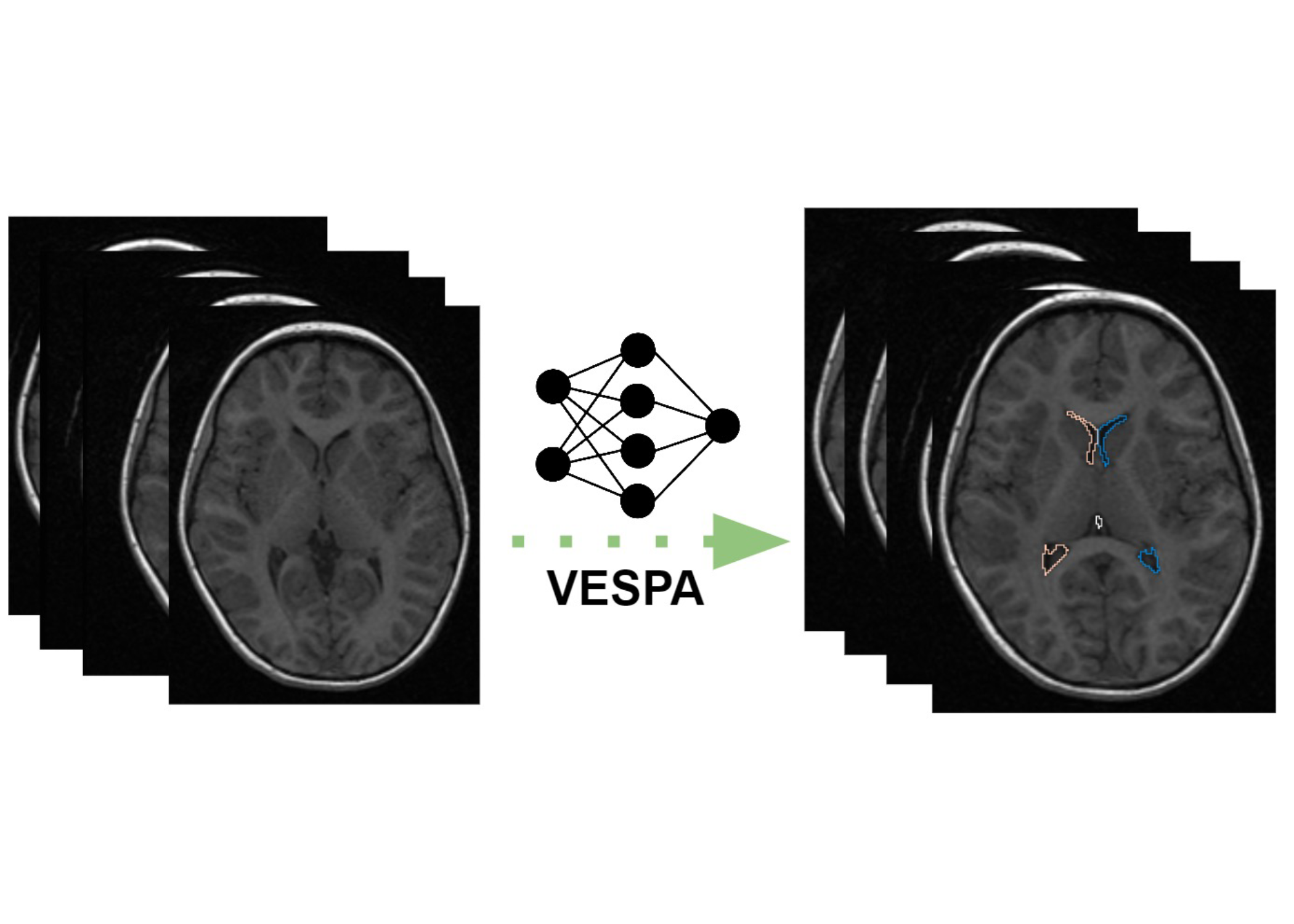
Dr. Birra Taha (Neurosurgery), Assistant Professor Ju Sun (Computer Science and Engineering; MSI PI), and Professor Daniel Guillaume (Neurosurgery) are working on a project called “Understanding Cerebral Pediatric Ventricular Growth Through Magnetic Resonance Imaging,” that seeks to utilize open and closed source data (M Health Fairview University of Minnesota Medical Center) to train a machine learning algorithm to segment the individual ventricular spaces in pediatric patients. This proposed automated method is called VESPA: VEntricular Segmentation in PediAtrics. Using ground truth segmentations obtained from multiple expert segmenters, the team hopes to train a fast, pathology-invariant neural network to segment the pediatric ventricular system (by compartment). Once trained, the model could be employed on open pediatric MRI datasets and eventually into local, closed-source data. The team hopes to publish these results in a free and easy-to-use container via GitHub, without any need for paid software.
This project recently received a Research Computing Seed Grant. Seed Grant funds are intended to promote, catalyze, accelerate and advance U of M-based informatics research, so that U of M faculty and staff are well prepared to compete for longer term external funding opportunities.
As of September 2023, the RC Seed Grant programs have been revised into the DSI Seed Grant programs. DSI Seed Grants include many of the same goals as the old program, with a new emphasis on data science. Complete information about DSI Seed Grants, including application deadlines, can be found on the RC website.