Diseases caused by plant pathogens reduce the global food crop by 40% annually. However, the bacteria and fungi that live in the soil (soil microbes) can produce an astounding diversity of antibiotic compounds that help crops and plants combat and suppress pathogens. In contrast to chemical-based pesticides, this antibiotic-mediated disease suppression by soil microbes presents a promising tool towards more sustainable disease control and yield stability, however, we still don’t understand much about the factors that influence the soil microbiome and antibiotic production. Particularly in agricultural contexts, we lack insight into the ways in which long-term farm practices, such as heavy fertilizer inputs, may alter the abundance and expression of antibiotics within the soil-microbiome. Thus, our ability to develop and manage disease-suppressive soils has remained severely limited.
In order to meet this need, PhD candidate Molly Kuhs (Ecology, Evolution, and Behavior) worked on a project called Does Long-Term Soil Nutrient Management Alter the Disease-Suppressive Capacity of Soil Microbes? as a UMII-MnDRIVE PhD Graduate Assistant. The goal of the project was to develop insight into how long-term agricultural practices, such as long-term nutrient inputs, influence the functional capacities of soil microbiomes. Critically, plant pathogens cause up to 40% food-crop losses annually and antibiotic mediated disease suppression by soil microbes presents a promising tool toward sustainable management and yield stability. The project focused on the role of antibiotics in mediating the plant disease-suppressive capacity of indigenous populations of soil microbes. Ongoing research combines a combination of genomic and phenotypic analyses to map responses among soil microbes under distinct nutrient management histories to test the hypothesis that long-term nutrient enrichment results in changes in inhibitory capacity, and thus disease suppressive potential of indigenous microbial populations.
Key findings of this project thus far include:
- Long-term nutrient amendment was correlated with significantly fewer inhibitory interactions among all soil microbial community members (25%) compared with nonamended community (47% of all interactions).
- Similarity in population-level inhibitory phenotypes between isolates is not correlated with 16S phylogenetic distance. That is, closely related individuals frequently did not share the same inhibitory phenotype.
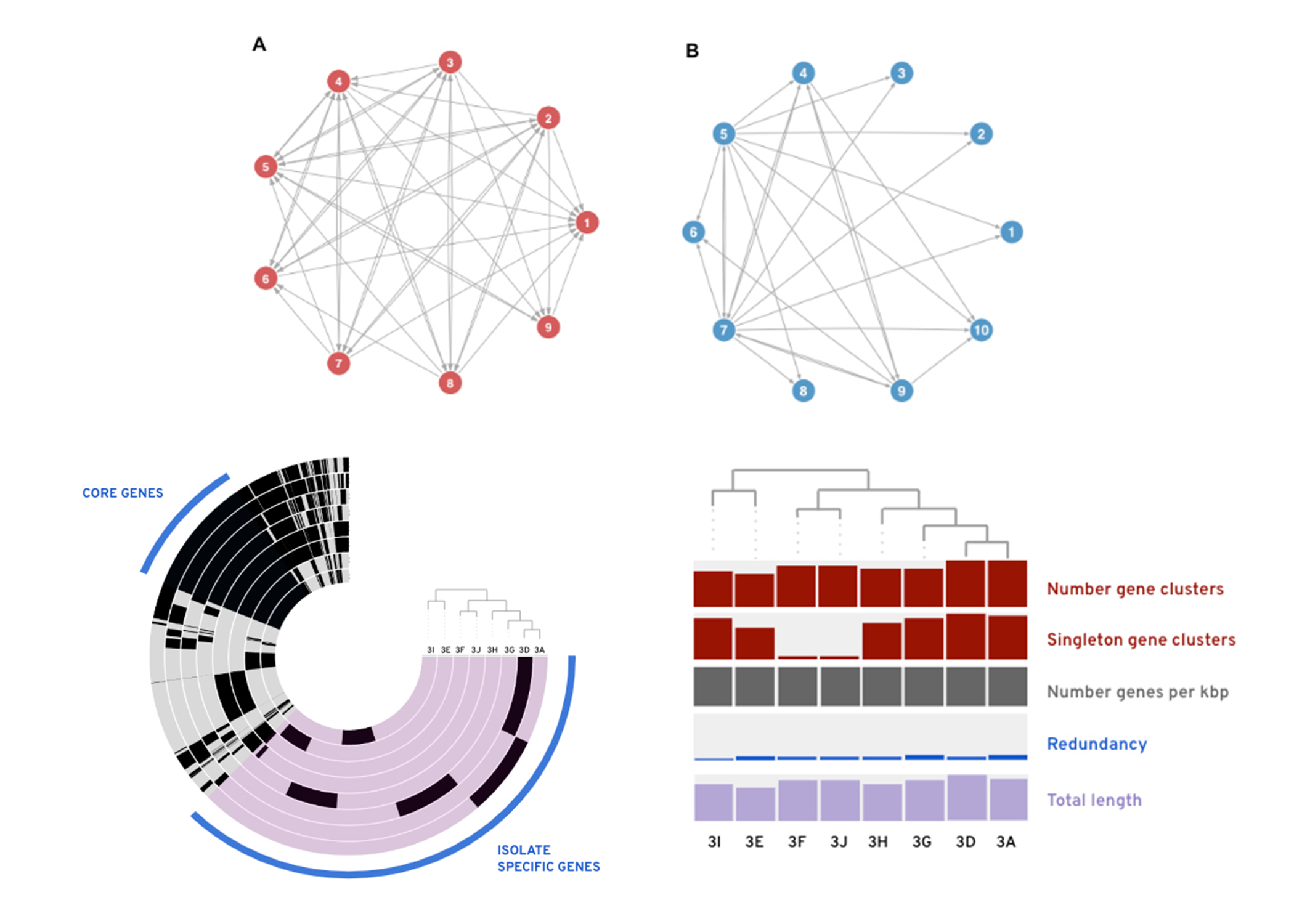
- Preliminary whole-genome analyses suggests that within the non-amended population, isolates have similar numbers of gene clusters beyond the set of core genes. Intriguingly, the number of unique, isolate-specific gene clusters varied across the population.
The next step in this project will be to analyze whole-genome sequences on amended isolates to compare genome size, core gene clusters, and isolate-specific antibiotic biosynthetic clusters to determine if long-term nutrient history alters the genomic structure of soil communities.
These findings suggest that the soil microbial population from long-term nutrient amendment displays significantly reduced inhibitory interactions across the soil population. Thus, long-term fertilization practices could result in significant reductions in soil-borne disease suppression and increased vulnerability of crop systems to disease. Continuing work will compare similarity and structure of antibiotic gene clusters between these two populations utilizing whole-genome sequences. Together these data will aid in optimization of the disease suppressive potential of indigenous soil microbiomes and provide context for developing novel agricultural disease management strategies.
Ms. Kuhs did this research under the supervision of Professor Linda Kinkel (Plant Pathology; MSI PI) and used MSI resources for bacteria identification and phylogeny building.
Preliminary findings from this project were presented in a poster at the 2022 Ecological Society of America’s (ESA) annual meeting in Montreal, Canada in August of 2022.
The UMII MnDRIVE Graduate Assistantship program supported U of M PhD candidates pursuing research at the intersection of informatics and any of the five MnDRIVE areas:
- Robotics
- Global Food
- Environment
- Brain Conditions
- Cancer Clinical Trials
This project was part of the Global Food MnDRIVE area.
The UMII program was converted to the Data Science Initiative-MnDRIVE Graduate Assistantship program in the fall of 2023. Research supported by the program is at the intersection of data science and the five MnDRIVE areas. The most recent Assistantships were announced in January 2024. The application period for the next awards will be announced in Fall 2024.
Image description:
Top: Within-population inhibitory interaction networks for long-term A) non-amended community, and B) amended community. Each node represents an individual isolate and connections indicating inhibition between community members with the arrow pointing from inhibitor to inhibited. 47% of total interactions in non-amended populations result in inhibition while only 25% of total interactions are inhibitory in long-term amended population.
Bottom: Preliminary pangenomic analysis of a subset of isolates originating from the nonamended community. Left: Whole genome mapping with blue highlight indicating core genes shared across all isolates, and purple region indicating gene-clusters unique to a single isolate in the population. Right: Whole-genome characteristics with bars are ordered by genetic similarity.