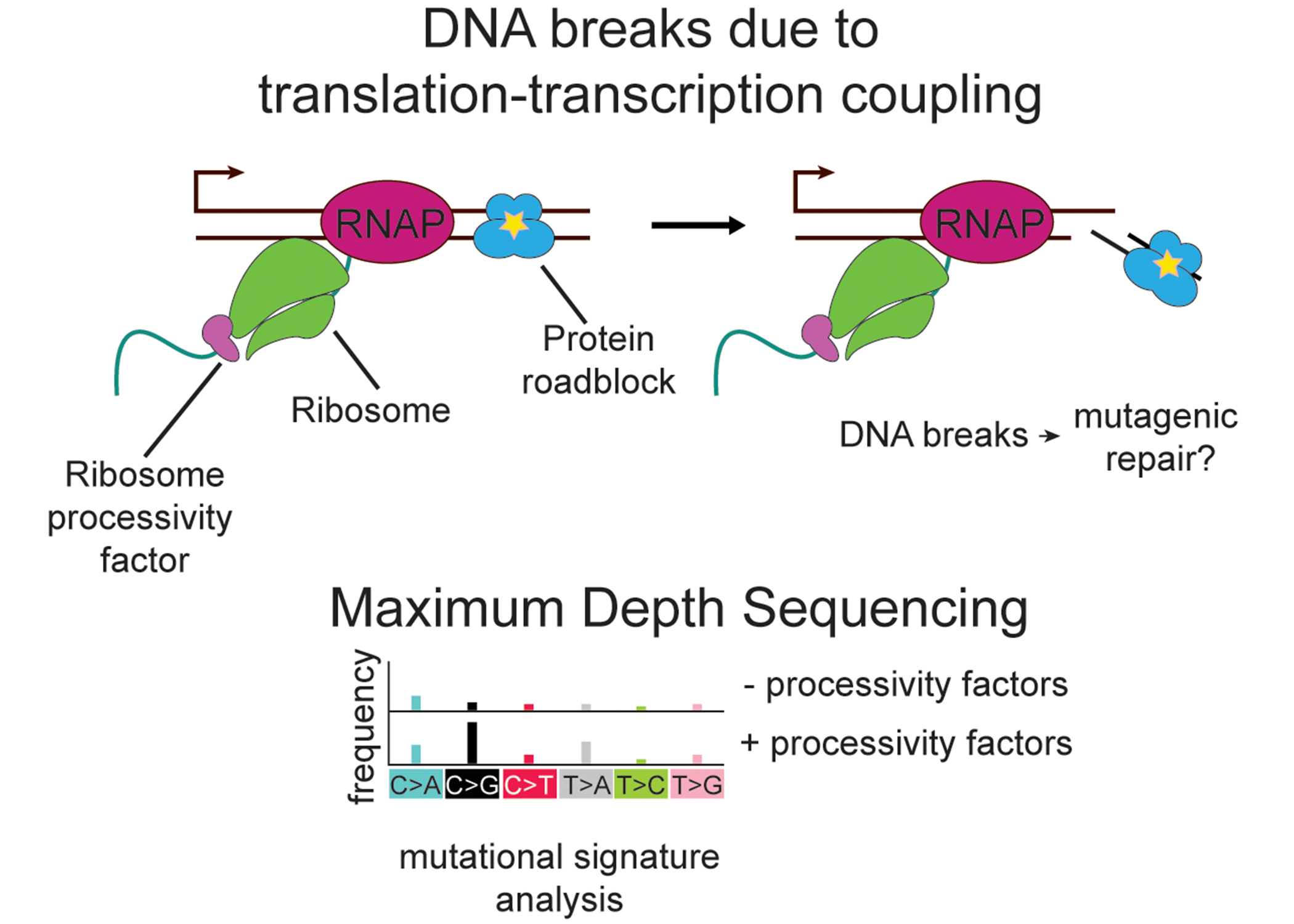
Antibiotic resistance is a global public health crisis. Fluoroquinolones are one of the most successful classes of antibiotics to date and are an incredibly important tool for treating infections. However, resistance to fluoroquinolones has become increasingly problematic. There is an urgent need for new antimicrobial compounds or compounds that can potentiate the action of those currently in use. Therefore, it is crucial that we understand the molecular mechanisms that lead to antibiotic resistance development in bacteria.
All living things require careful coordination of DNA replication, transcription, andtranslation. In bacteria, these three processes are spatially and temporally coupled.Understanding how this is orchestrated is crucial. Translation of most nascent mRNAs begins soon after the ribosomal binding site (RBS) emerges from the RNA exit channel of RNA polymerase (RNAP). Importantly, translation-transcription coupling promotes RNAP translocation and transcription elongation. Translocating RNAPs can collide with tightly bound DNA gyrase complexes that have been covalently crosslinked by fluoroquinolone antibiotics, resulting in DNA damage. This DNA damage is thought to be the major bactericidal mechanism of fluoroquinolone antibiotics. Furthermore, induction of DNA damage can lead to mutagenic repair processes, resulting in increased evolution of antimicrobial resistance. Therefore, it is important to understand how translation impacts RNAP translocation and subsequently genome integrity. Using the Gram-positive model organism Bacillus subtilis, these researchers have found that increasing ribosome stalling on nascent mRNAs drastically reduces sensitivity of cells fluoroquinolone treatment. Furthermore, they have found that in cells lackinghomologous DNA repair pathways are less sensitive to fluroquinolone treatment if their ribosome mobility is impaired. They therefore hypothesize that ribosome movement on nascent mRNA modulates RNAP mobility resulting in reduced DNA damage and/or improved DNA repair during fluoroquinolone treatment.
Assistant Professor Kevin Lang (Veterinary and Biomedical Sciences; MSI PI) is working on a project called “Molecular mechanisms of evolution in bacteria,” that seeks to better understand the bactericidal action of fluoroquinolone antibiotics, and its impact on antibiotic resistance development by studying translation-transcription coupling in bacteria. They will test this hypothesis with the following objectives:
- Determine the impact of translation-transcription coupling on DNA breaks
during fluoroquinolone treatment. - Determine the impact of translation-transcription coupling on mutation rates.
Understanding the mechanisms that lead to resistance will aid in the development of strategies to reduce the frequency and dissemination of resistant pathogens. Furthermore, understanding the mechanisms of antimicrobial action will help determine novel targets for the development of potentiating compounds that will increase the efficacy of antibiotics currently in use.
This project recently received a Research Computing Seed Grant. Seed Grant funds are intended to promote, catalyze, accelerate and advance U of M-based informatics research in areas related to the MnDRIVE initiative, so that UMN faculty and staff are well prepared to compete for longer term external funding opportunities. This Seed Grant falls under the Global Food research area of the MnDRIVE initiative.
As of September 2023, the RC Seed Grant programs have been revised into the DSI Seed Grant programs. DSI Seed Grants include many of the same goals as the old program, with a new emphasis on data science. Complete information about DSI Seed Grants, including application deadlines, can be found on the RC website.